Meller lab team new paper in Advanced Materials journal
Full-Length Single Protein Molecules Tracking and Counting in thin Silicon Channels
In this paper, we introduce a single-molecule method for parallel protein separation and tracking, yielding multi-dimensional and are electrophoretically separated by their mass/charge in custom-designed thin silicon channel with subwavelength height. This approach allows us to analyse thousands of individual proteins within a few minutes by tracking their motion during the migration. We demonstrate the power of the method by quantifying a cytokine panel for host-response discrimination between viral and bacterial infections.
Neeraj Soni won the best presentation award at Single Molecule Biophysics Conference, France 2022
Neeraj Soni, a Ph.D. student in our group, won the best presentation award at Single Molecule Biophysics Conference, France 2022.
His presentation title : "Denatured protein translocations using sub 5 nm Solid-state nanopores”.
Proteins are the main constituents of all biological cells and are responsible for most of the cellular functions. Many of the current disease-specific biomarkers are proteins or modified versions of proteins hence creating a need for sensitive yet simple methods for protein identifications in clinical diagnostics applications. Herein, we developed a highly sensitive approach for identifying wide range lengths SDS denatured protein complexes. Given the simplicity and eradication of the tailor-made necessities, this assay holds the full potential to sequence the protein at a single-molecule level in the future.
Way to go Neeraj!
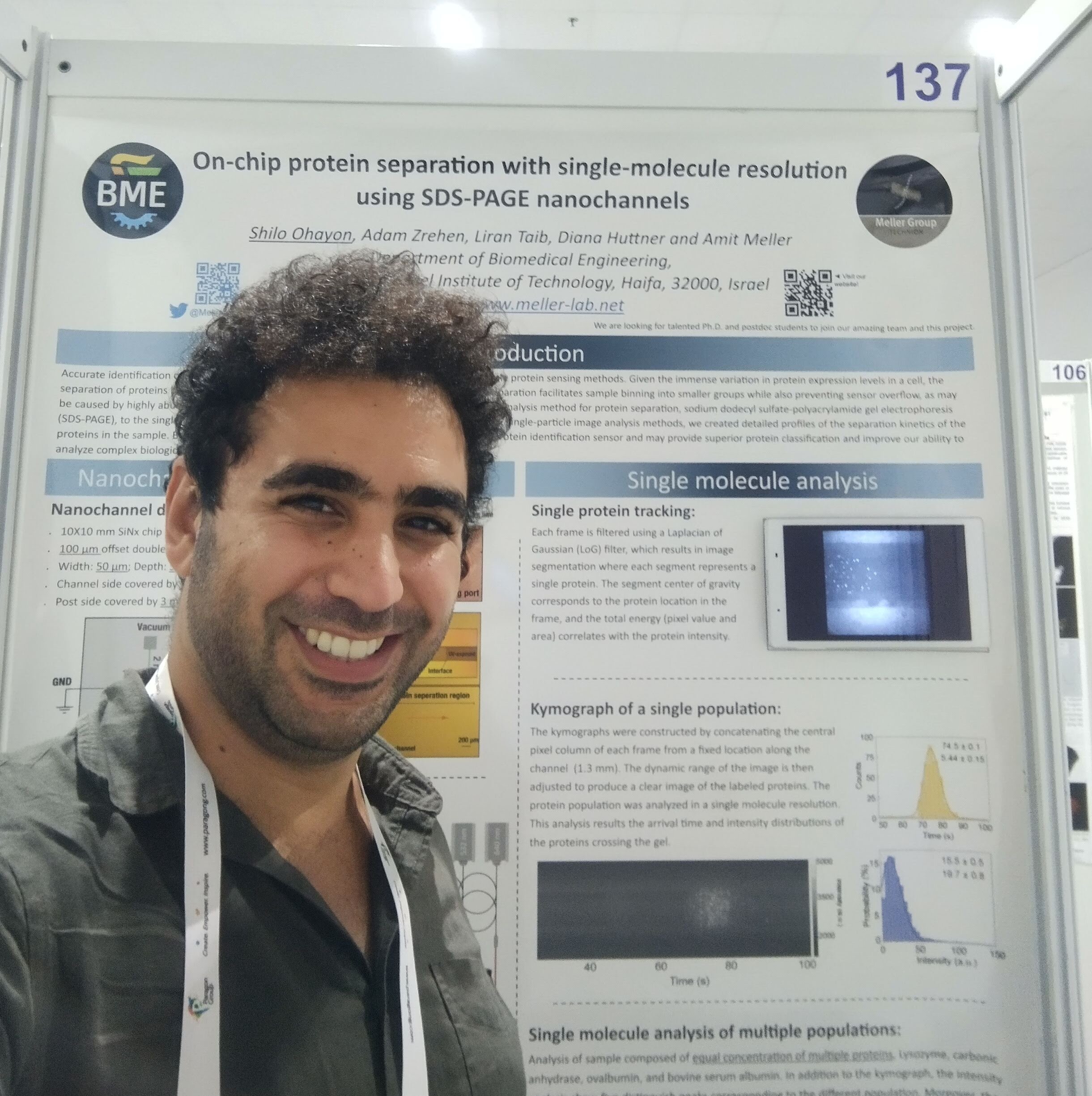
Congratulation to Shilo Ohayon!
Shilo Ohayon, a Ph.D. student in our group, won the best poster presentation at the NanoIL 2021.
His poster title : "On-chip protein separation with single-molecule resolution using SDS-PAGE nanochannels”.
The research's main aim is to identify and quantify proteins at a single molecule level using nanochannels. The use of nanochannel allows real-time tracking of the translocating proteins by a computer vision algorithm. The data obtained from each protein enables the identification of the protein and, by that, the whole sample. In the future, this technology could manifest faster and more efficient identification of proteins in biological and clinical samples and eventually will help in more accurate diagnostics.
The lab perspective paper on the emerging field of single-molecule protein identification is out on Nature Methods
Single-cell profiling methods have had a profound impact on the understanding of cellular heterogeneity. While genomes and transcriptomes can be explored at the single-cell level, single-cell profiling of proteomes is not yet established. Here we describe new single-molecule protein sequencing and identification technologies alongside innovations in mass spectrometry that will eventually enable broad sequence coverage in single-cell profiling. These technologies will in turn facilitate biological discovery and open new avenues for ultrasensitive disease diagnostics.
Congratulations to Shilo Ohayon for winning the Jacobs Excellence Reward for the best published paper.
Shilo Ohayon, a Ph.D. student in our lab, won the Jacobs Excellence Reward for the best published paper for the current academic year 2020-2021.
Shilo’s Paper : “On‑chip protein separation with single‑molecule resolution”. Sci Rep 2020, 10-15313–12.
Way to go Shilo!
Congratulations to Nitinun for the publication of her Chem Society Review paper
Proteins are the structural elements and machinery of cells responsible for a functioning biological architecture and homeostasis. Advances in nanotechnology are catalyzing key breakthroughs in many areas, including the analysis and study of proteins at the single-molecule level. Nanopore sensing is at the forefront of this revolution. This tutorial review, published on October 17, 2018, provides readers a guidebook and reference for detecting and characterizing proteins at the single-molecule level using nanopores. Specifically, the review describes the key materials, nanoscale features, and design requirements of nanopores. It also discusses general design requirements as well as details on the analysis of protein translocation. Finally, the article provides the background necessary to understand current research trends and to encourage the identification of new biomedical applications for protein sensing using nanopores.